Our research focuses on the investigation of targeting mechanisms that recruit select mRNA transcripts to specific subcellular localizations and aims to understand their contribution to disease. We develop live single-molecule imaging tools and imaging-based spatial transcriptomics assays to combine them with novel 3D organoid model systems.
Pioneer studies have quantified global biases in RNA localization and subcellular differences in translation efficiency that allow local regulation of gene expression (incl. Voigt et al., 2017). Yet most have failed to dissect the molecular mechanisms that target specific transcripts. In the past, this was due to a lack of single-molecule methods as well as to the inherent heterogeneity of RNA localization patterns observed in individual cultured cells in the absence of a clear polarization axis.
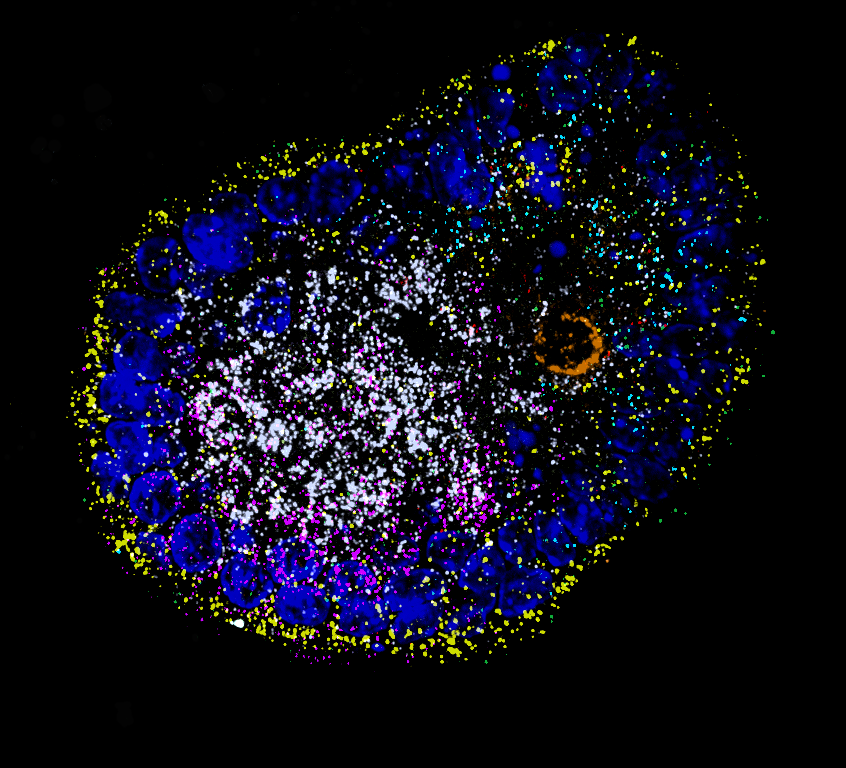
To remedy this, we employ clearly polarized organoid models that allow us to investigate RNA localization on the single-molecule, single-cell, as well as tissue level – all in a single multi-scale experimental set-up.
Dynamics and mechanisms underlying RNA localization
To dissect RNA targeting mechanisms and assess how they regulate gene expression locally, we quantify recruitment dynamics, translational efficiency and turnover of individual mRNA transcripts in different subcellular localizations using live single-RNA imaging approaches.
We have established reporter transcripts that are stably expressed in murine intestinal organoid lines engineered to allow live single-particle imaging.
To characterize transport complexes at the molecular level, we assess their mode of transport using single-particle co-localization and diffusion analysis. In addition, we aim to identify the trans-acting factors that recruit RNAs to their target sites through a combination of in-vitro biochemistry approaches.
Global RNA localization patterns
In order to map RNA localization patterns on a transcriptome-wide scale, we take a holistic and unbiased approach to identify novel cis-acting elements and correlate RNA localization with physiological function:
We use imaging-based spatial transcriptomics to systematically record localization patterns of many different endogenous transcripts in parallel and employ external stimuli to quantify changes in RNA localization biases, which will allow us to identify novel targeting motifs. For validation of our findings, we use single-cell genomics in collaboration with Treutlein lab, ETH Zurich DBSSE.
Our model systems are primarily murine intestinal organoids, which robustly recapitulate RNA localization biases that were previously described in literature. As well as more recently human iPSC-derived cerebral cysts and brain organoids that we have started to characterize as a multicellular model system to study RNA targeting in neuronal processes and at synapses.
XBP1 mRNA splicing on the ER
The unfolded protein response (UPR) relies on the non-canonical splicing of XBP1 mRNA on the surface of the endoplasmic reticulum (ER). The molecular switch that initiates splicing is the oligomerization of the ER stress sensor and UPR endonuclease IRE1α (inositol-requiring enzyme 1 alpha).
While IRE1α can form large clusters that have been proposed to function as XBP1 processing centers on the ER, the actual oligomeric state of active IRE1α complexes as well as the targeting mechanism that recruits XBP1 to IRE1α oligomers remains unknown.
We have developed a single-molecule imaging approach that can monitor the recruitment of individual XBP1 transcripts to the ER surface, and used it to visualize how IRE1α-catalyzed splicing mobilizes XBP1 mRNA from the ER membrane in response to ER stress. Our current work focuses on quantifying the dynamics of the transient XBP1-IRE1α interactions on the ER surface.
Gene expression dynamics
during early human brain organoid development
Cerebral organoids have revolutionized the study of human brain development because they model the different stages through which neural tissue transitions in vitro.
Single-cell genomics methods have provided very detailed insights into which genes are expressed in which cell types at the various stages of neurogenesis, yet little is known about how the timing and fluctuation of gene expression influences cell fate decisions and morphogenesis.
To address these questions, we have developed an imaging-based approach that allows us to monitor gene expression in real time and correlate it with cell type- and state specific information. We investigate the dynamics of gene transcription in the context of early neural development and correlate the obtained data with measurements of RNA and protein content in fixed tissues.
Ultimately, our approach will allow us to resolve how small differences in the timing, localization and frequency of gene expression result in the formation of diverse specialized cell types and tissue regions.